Bioinformatics and Systems Biology
“Bridging the Gap Between Molecular Science and Improved Health Outcomes”
ICBI develops and applies computational pipelines for analysis of molecular profiling data from high-throughput genome wide technologies such as Next Generation Sequencing, gene and microRNA expression, DNA copy number, proteomics, and metabolomics. We apply a wide range of statistical and computational approaches such as machine learning, regression and correlation analysis for integration of molecular data with clinical attributes, correlating the results from high-throughput molecular platforms, patient characteristics, and outcomes to improve public health. As such, we collaborate with numerous experts in basic and translational biomedical sciences, cancer research, regulatory science, health educators, and other data scientists.
Current Projects
Data Types: Whole Genome-seq , RNA-seq, Small RNA-seq and Whole Exome-seq
Our team has developed several pipelines for NGS data processing and analysis utilizing high-performance cloud computing platforms such as AWS and Globus Genomics. We are currently focusing on variant analysis for Whole Genome sequencing and gene expression analysis based on RNA-seq data in a translational research setting. Several peer reviewed papers include analysis of NGS data in cancer related projects.
Multi-Omics Molecular Profiling of Human Cancers
A number of collaborative projects is underway that are dealing with analysis of multi-omics profiling data for breast cancer, colorectal cancer, pancreatic cancer, liver cancer and other diseases. The studies include on-going projects with several principal investigators at Lombardi Comprehensive Cancer Center as well as National Children’s Hospital , and Howard University. To date we have published nearly 20 peer reviews articles reporting differential expression analysis of gene expression, microRNA expression, DNA copy number, and metabolomics in human cancers.
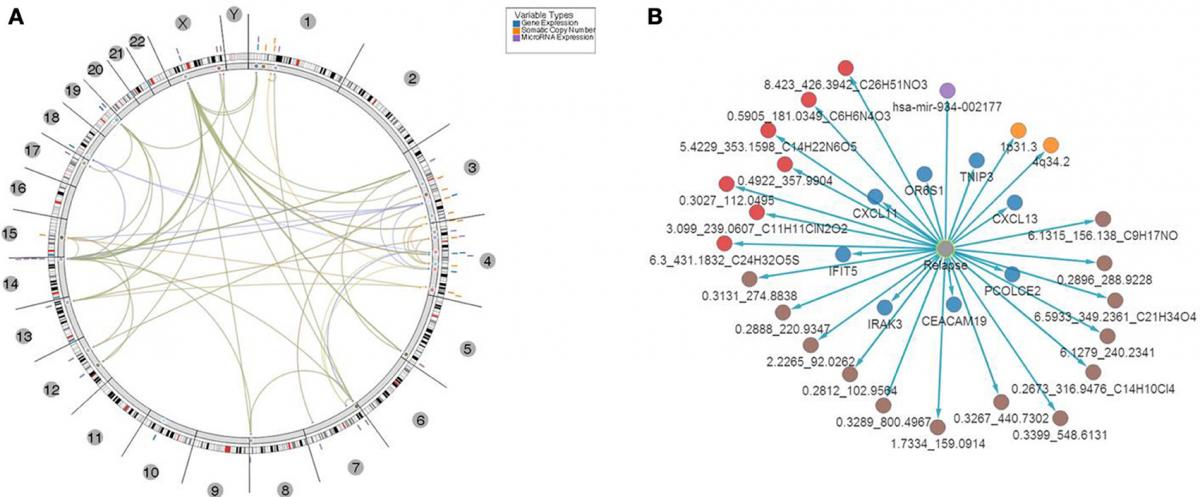
CRC Results Circos Plot Plus Network
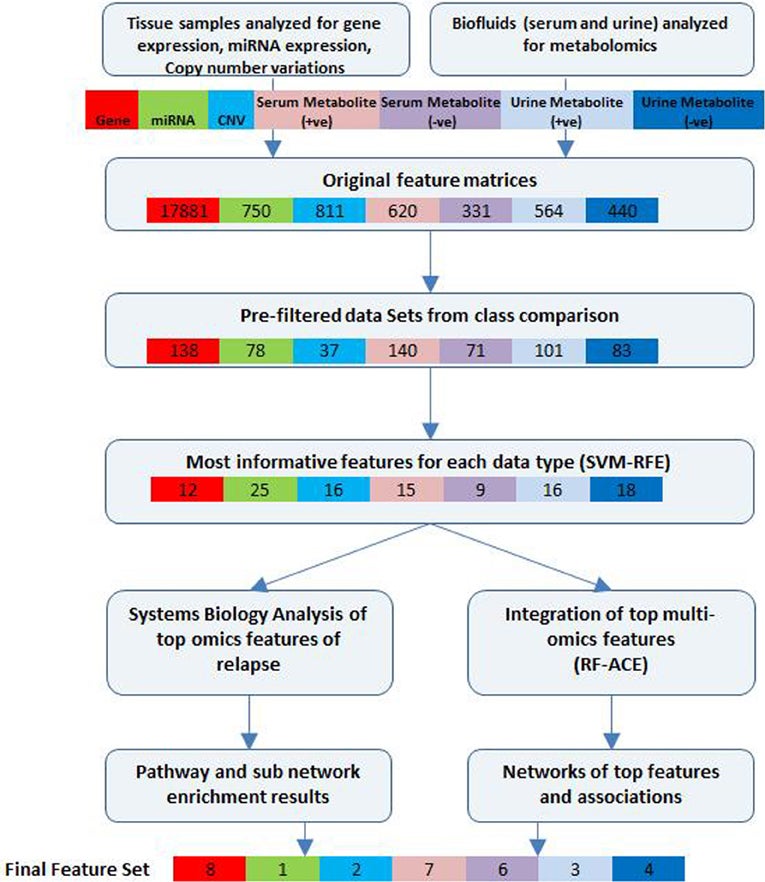
Intergrative Analysis Workflow
Metabolomics
Metabolites are small molecules. They can be measured in a variety of biospecimens, including blood, urine, and tissue. As they are intermediates and products of metabolism, they reflect both genetic and environmental factors and have recently been the focus of much research in a variety of disease areas. Metabolomics projects require using and developing special pre-processing and annotation approaches as well as building scientifically relevant models. Members of our group have collaborated on studies of metabolomics in cancer, rare diseases, and chronic disorders.
Predictive Modeling and Personalized Medicine
In collaboration with the personalized medicine team at ICBI, we are analyzing several datasets related to personalized medicine. Our focus is to look at commercially-available tumor profiling assays and to build improved predictive models for recommending specific therapies for advanced cancer patients.
Data Integration and Systems Biology Analysis
Our team is working with computational methodologies for integration of multiple molecular data types together with clinical outcome and/or drug response information utilizing machine learning algorithms such as SVM-RFE, Random Forest, Elastic Net, Lasso, and others. We are also using various systems biology analysis tools to provide additional level of integration utilizing prior biological knowledge to map the data to pathways and interaction networks in order to get biological insights and biological interpretation of the high throughput genomes wide profiling data.
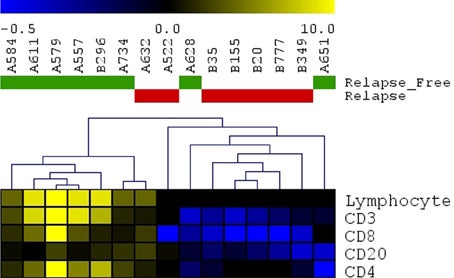
Heatmap Infiltr Lymphocytes
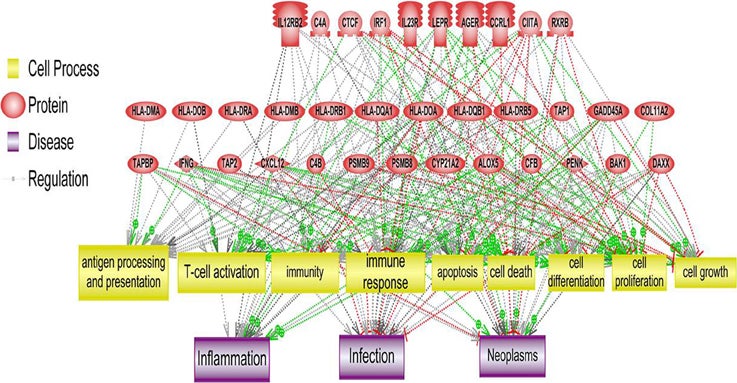
PS Downstream Target Analysis
Collaborations
“Integration of ER-related signaling in breast cancer”
ICBI is involved in a massive systems biology effort, rewarded by a U54 grant (PI: Dr. Robert Clarke, Georgetown University). Our work was presented at several conferences and two papers are currently in preparation.
“Biomarker discovery and validation in a Duchenne dystrophy natural history study”
Our team is collaborating in an effort to find metabolomic and proteomic markers of progression for Duchenne Muscular Dystrophy, a rare degenerative disease (PI: Dr. Craig McDonald, UC Davis). Our work was presented at several conferences and two papers are currently in preparation.
“Metabolomics in Gulf War Illness”
We are collaborating with Dr. James Baraniuk (Georgetown University) to identify metabolic biomarkers for Gulf War Illness, which affects many veterans who served in the 1991 Gulf War.
“Targeting Triple Negative Breast Cancer in African-American Women”
This collaboration is funded by the FDA with Dr. Luciane Cavalli from Lombardi Cancer Center to identify novel biomarkers for triple negative breast cancer in several minority populations using DNA copy number and microRNA profiling.
Publications Resulting from Work in this Research Platforms
Pathak R, Cheema AK, Boca SM, Krager KJ, Hauer-Jensen M, Aykin-Burns N. “Modulation of radiation response by the tetrahydrobiopterin pathway.” Antioxidants, 2015, 4(1), 68-81
Guertin KA, Loftfield E, Boca SM, Sampson JN, Moore SC, Xiao Q, Huang WY, Xiong X, Freedman ND, Cross AJ, Sinha R. “Serum biomarkers of habitual coffee consumption may provide insight into the mechanism underlying the association between coffee consumption and colorectal cancer.” American Journal of Clinical Nutrition, in press (2015).
Subha Madhavan, Yuriy Gusev, Salendra Singh and Rebecca B Riggins. ERRgamma target genes are poor prognostic factors in Tamoxifen-treated breast cancer. Journal of Experimental & Clinical Cancer Research. In Press (2015)
Krithika Bhuvaneshwar, Dinanath Sulakhe, Robinder Gauba, Alex Rodriguez, Utpal Madduri Ravi, Dave, Lukasz Lacinski, Ian Foster, Yuriy Gusev, Subha Madhavan. A case study for cloud based high throughput analysis of NGS data using the Globus Genomics system. Computational and Structural Biotechnology Journal, 11/2014
βII-spectrin (SPTBN1) suppresses progression of hepatocellular carcinoma and wnt signaling via regulation of wnt inhibitor kallistatin. Hepatology. 10/2014; DOI: 10.1002/hep.27558
Transcriptional Regulation of STAT3 by SPTBN1 and SMAD3 in HCC Through cAMP-Response Element-Binding Proteins ATF3 and CREB2. Carcinogenesis. 08/2014; DOI: 10.1093/carcin/bgu163
Costello,J.C., et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nature Biotechnology (2014); 06/2014; DOI: 10.1038/nbt.2877
Torresan, C. et al. Increased copy number of the DLX4 homeobox gene in breast axillary lymph node metastasis. Cancer Genetics 2014, DOI: 10.1016/j.cancergen.2014.04.007
S Madhavan, R Gauba, R Clarke, Y Gusev. Integrative Analysis Workflow for Untargeted Metabolomics in Translational Research. Metabolomics, 2014 4 (130), 2153
S Madhavan, Y Gusev et al. Genome-wide multi-omics profiling of colorectal cancer identifies immune determinants strongly associated with relapse. Frontiers in Cancer Genetics. 2013, 4: 236
Elgamal OA, Park J-K, Gusev Y, Azevedo-Pouly ACP, Jiang J, et al. Tumor Suppressive Function of mir-205 in Breast Cancer Is Linked to HMGB3 Regulation. PLoS ONE, 2013, 8(10): e76402.
Gusev Y, Riggins RB., Krithika Bhuvaneshwar, Robinder Gauba, Laura Sheahan, Robert Clarke, and Subha Madhavan. In silico discovery of mitosis regulation networks associated with early distant metastases in estrogen receptor positive breast cancers. Cancer Informatics, 2013;12:31-51
C Albanese, OC Rodriguez, J VanMeter, ST Fricke, BR Rood, YC Lee, SS Wang, S Madhavan, Y Gusev, EF Petricoin III, Y Wang. Preclinical Magnetic Resonance Imaging and Systems Biology in Cancer Research: Current Applications and Challenges. Preclinical Magnetic Resonance Imaging and Systems Biology in Cancer Research: Current Applications and Challenges. The American Journal of Pathology, December 4, 2012
R Gauba, TG Natarajan, L Song, K Bhuvaneshwar, S Madhavan, Y Gusev. Metabolomic and exome sequence analysis reveal novel molecular signatures associated with colorectal cancer relapse. BMC Proceedings 6 (Suppl 6), P41, 2012
Madhavan S, Y Gusev, MA Harris, DM Tanenbaum, R Gauba, K Bhuvaneshwar, A Shinohara, K Rosso, L A Carabet, L Song, R B Riggins, S Dakshanamurthy, Y Wang, S W Byers, R Clarke, L M Weiner. G-CODE: enabling systems medicine through innovative informatics. Genome Biology 12 (Suppl 1):P38, September 2011
Madhavan S., Gusev Y., Harris M. et al. G-DOC®: A Systems Medicine Platform for Personalized Oncology. Neoplasia. 2011 Sep;13(9):771-83
Mark A Semenuk, Timothy D Veenstra, Gradimir Georgevich, Mark Rochman, Yuriy Gusev, Elizabeth J Haanes, Branka M Bogunovic, Michael Lebowitz, Nikolai Daraselia, Elena I Schwartz. Translating Cancer Biomarker Discoveries to Clinical Tests: What should be Considered? Recent Patents on Biomarkers 2011, 1: 222-240
Park JK, Henry JC, Jiang J, Esau C, Gusev Y. et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011 Feb 14. [Epub ahead of print]
