Technology and Health IT
“Creating Software and Tools to Enable Knowledge Discovery and Clinical Decision Making.”
The research information technology group at ICBI develops innovative scientific software to enable translational research. Our projects include muti-omics data analysis, vaccine safety research, clinical data analysis, high definition data visualization, natural language processing, and mobile application development.
The Health Information Technology/ Software and Engineering team functions at the intersection of information science, computer science, and health care. The team manages and supports computing and communications technology applied to healthcare, health education, and research as well as the use and sharing of information within the institutions in support of our investigators. The team develops custom software applications, tools, and databases and manages the resources, devices, and methods required for optimizing the acquisition, storage, retrieval, analysis and use of information. It also manages dedicated servers, virtual machines both within the Georgetown premises and in the cloud, configured with strict security measures, restricted backup devices and capable of collecting, storing, analyzing, and distributing big data while facilitating collaboration with laboratories across the world. Specialized software, development packages and expertise are available for decision support, molecular modeling, processing, advanced statistical data analysis, knowledge management, mathematics, data mining and big data processing. The computing capabilities ICBI manages, include the latest technologies and secure environments for conducting data collection to do research and education or clinical care. The center is also leading advanced data visualization infrastructure within Georgetown University Medical Center (GUMC), a technology that allows researchers to visualize large data sets in high fidelity, displaying over 16 million pixels of data in a single view.
Current Projects
G-DOC
The Georgetown Database of Cancer Plus other diseases (G-DOC Plus) is a precision medicine platform containing molecular and clinical data from thousands of patients and cell lines, along with tools for analysis and data visualization. The platform enables the integrative analysis of multiple data types to understand disease mechanisms. G-DOC Plus has three overlapping entry points for the user based on their interests: 1) Personalized Medicine, 2) Translational Research, and 3) Population Genetics.
View G-DOC Tutorials and webinar recordings.
G-VISR
Application to analyze and curate information about the molecular basis of vaccine safety.
MedTurk
Helps researchers automate the process of extracting information from clinical notes and data from electronic health records.
Biospecimen Portal
This portal integrates data about biospecimens from the Lombardi Shared Resources allowing investigators to identify patients and samples for research projects.
Data Visualization Lab
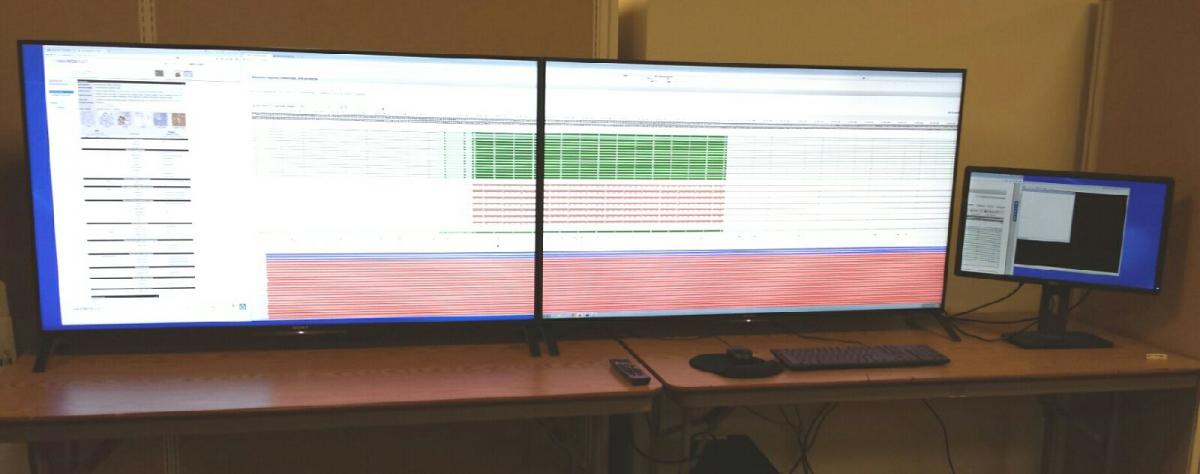
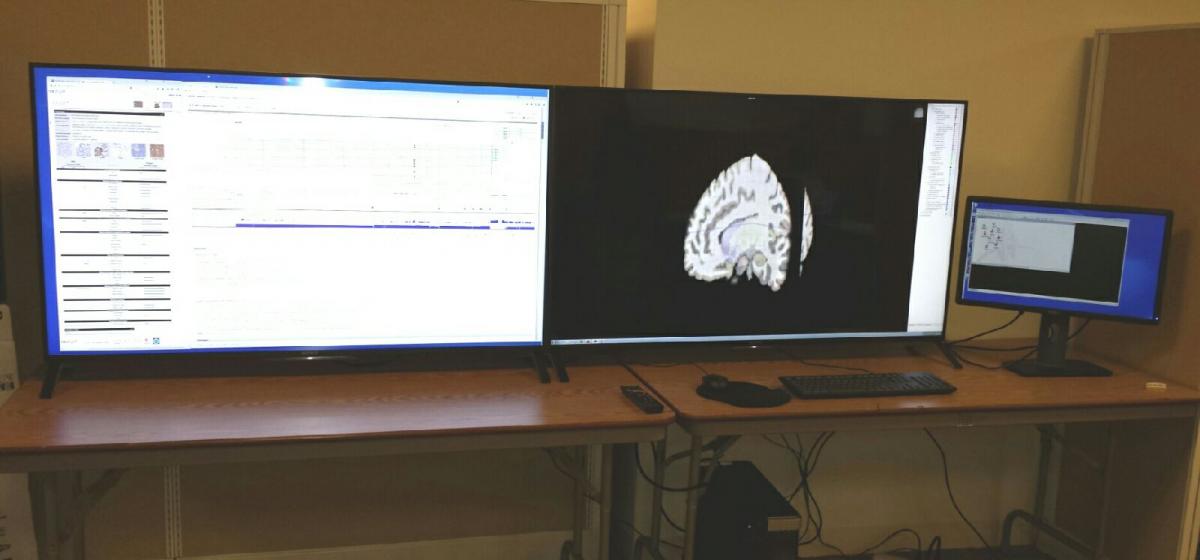
The ICBI visualization system was created through an NVIDIA corporation hardware donation grant and hardware contributed by the University Information Systems department. High definition visualization hardware for big data visual analytics. The visualization lab consists of 2 ultra high definition 55 inch display panels which are driven by NVIDIA Quadro K5000 high end video card. The system is capable of displaying 16 million mega pixels.
GWCAM Mobile Application
A mobile application to collect quality of life information from gulf war veterans.
Collaborations
Amazon Web Services
ICBI has received a grant from Amazon.com (3 cycles) to conduct research in high performance computing using AWS.
ICBI developed comprehensive processes and tracking and monitoring systems to manage requests and be able to track and measure key performance indicators.
GHUCCTS
The Georgetown-Howard Universities Center for Clinical and Translational Science (GHUCCTS) is a collaborative research center that includes two major universities and three affiliated hospital and research systems. GHUCCTS institutions include the Georgetown University Medical Center, Howard University, MedStar Health Research Institute, the Washington DC Veterans Affairs Medical Center (VAMC) and Oak Ridge National Laboratory. Patient Data Access: To facilitate clinical trial recruitment and identify patients for research studies across GHUCCTS institutions, we established a secure exchange architecture that enables intra- and inter-institutional clinical and translational research information sharing across boundaries; we also developed cohort discovery methodologies for querying electronic health records across our participating institutions.
For this purpose, we implemented a combination of tools that were configured according to GHUCCTS data governance policies. These tools include Explorys, a grid based population discovery application and the Integrating Biology and the Bedside (i2b2) software, an open source analytical tool. The combination of these systems will ensure that researchers at each of the GHUCCTS institutions will be able to easily identify potential clinical trial participants across institutions, while protecting the confidentiality and security of their health information.
The established tools and capabilities will also support the analysis and visualization of research and clinical data and enable investigators easy access to available populations across multiple institutions. The implementation entailed the design and development of software applications to consistently map data from multiple sources into a common repository, and to create a process and implement tools for facilitating secure information sharing. In our setting it is challenging to implement an integrated research data repository that includes all participating institutions’ data. Our final approach was to establish a searchable GHUCCTS research data repository complemented by a process to handle query submission to institutions and data sources which cannot be readily integrated into the GHUCCTS i2b2 repository.
- WebCAMP
- SharePoint
- i2b2, Explorys
- EHR systems
- Lab systems
- REDCap
Technology

The development of research infrastructure is the foundation of the Biomedical Informatics program at Georgetown and underlies the mission of integrating and making sense of enormous volumes of data being generated in both the lab and clinic. ICBI scientists and software engineers are working together to develop technologies that enable the integration of biomedical data with state-of-the-art tools through a multi-disciplinary and collaborative approach that drives translational research and clinical care.
Our primary areas of technology development involve:
