Services Vignettes
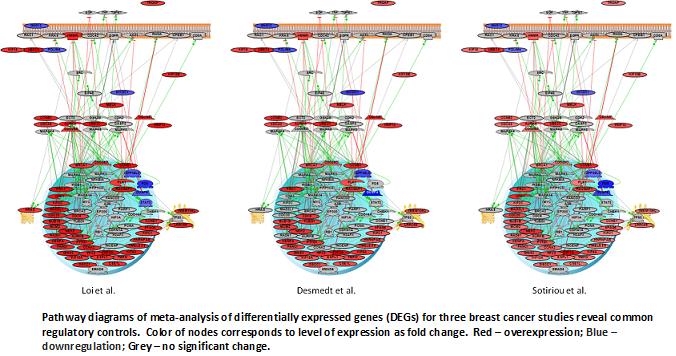
Meta-analysis of publically available breast cancer data
ICBI members have been working with Dr. Rebecca Riggins, a cancer biologist within Lombardi, and Dr. Bob Clarke, Dean for Research at GUMC on to analyze genomic data (microarray and exome-seq) from breast cancer cell lines. One project recently published in Cancer Informatics was to examine several differentially expressed genes across publicly available breast cancers studies to find genomic features leading to early metastasis in ER+ breast cancer. ICBI members used several bioinformatics analysis tools to both determine the common differentially expressed genes across studies and to do biological pathway analyses to discover common regulatory gene networks disrupted across the studies.
Genetic contributions to individual variability in drug response – a pharmacogenomics analysis
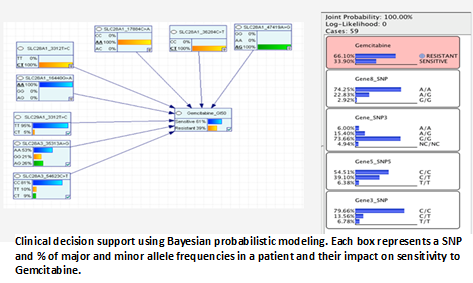
ICBI members are collaborating with Dr. John Deeken, an oncologist and cancer researcher within Lombardi, to develop methods to identify genes contributing to the variation in drug response among patients, by using genotype information from drug metabolizing enzymes and transporter genes (DMET genes). Employing the Affymetrix DMET platform, Dr. Deeken has generated genotyping data for all NCI 60 cell lines, which is a set of publically available human cancer cell lines derived from diverse tissues. Using this data ICBI is helping Dr. Deeken to study the effects of genomic variation on drug sensitivity, resistance, and mechanisms of drug action. We have integrated the NCI 60 genotyping data with other data sources including publically available data such as GI50 and gene expression data from sources like PharmGKB (http://www.pharmgkb.org), Drugbank (http://drugbank.ca) and the FDA Developmental Therapeutics Program (DTP) public website. Bioinformatics tools both existing and newly developed are being utilized to integrate the data from these disparate sources. Among the tools employed are Bayesian Belief Networks, which are being used to model the effect of DNA sequence variation on drug metabolism using a pathway centric approach.This endeavor should provide novel insights into both pre-existing and future drug efficacy and toxicology studies.
Support for clinical research data management
REDCap (Research Electronic Data Capture) is a secure, web-based applicationdeveloped at Vanderbilt University that supports centralized data collection for research studies. The ICBI group actively supports a locally hosted instance of REDCap for use by the Georgetown University research community. ICBI staff have helped dozens of investigators at GU set up RedCap to organize large clinical studies. One example is the Liver Transplant Database developed for the Georgetown Transplant Institute.
Common Data Elements (CDEs) and Case Report Forms (CRFs) can be easily implemented in REDCap using a flexible and user-friendly form builder to provide a standardized data entry portal for the study metadata. REDCap provides advanced features for data validation, enforcement of restricted vocabularies, electronic signature, file upload, record and study locking, and role based access. In addition, REDCap can compartmentalize study data to support simultaneous data collection from numerous multi-site clinical trials. REDCap provides a user-friendly display that marks completed forms with a green icon and incomplete forms with a red icon, allowing a user to quickly view which forms for a certain case are complete or incomplete. REDCap also provides the user with pre-configured forms library containing commonly used case report forms. Case report forms and common data elements developed by the clinical centers can be implemented in REDCap to provide a standardized data entry portal for the study metadata. REDCap also has robust functionality to de-identify data for distribution to the research community, if needed.
There are currently over 100 active projects in the Georgetown hosted REDCap instance. Training for REDCap is provided by members of the ICBI staff to interested researchers. Plans are underway to integrate de-identified data from REDcap into a standardized database to enable cohort discovery for future research needs at Lombardi and the medical center.
